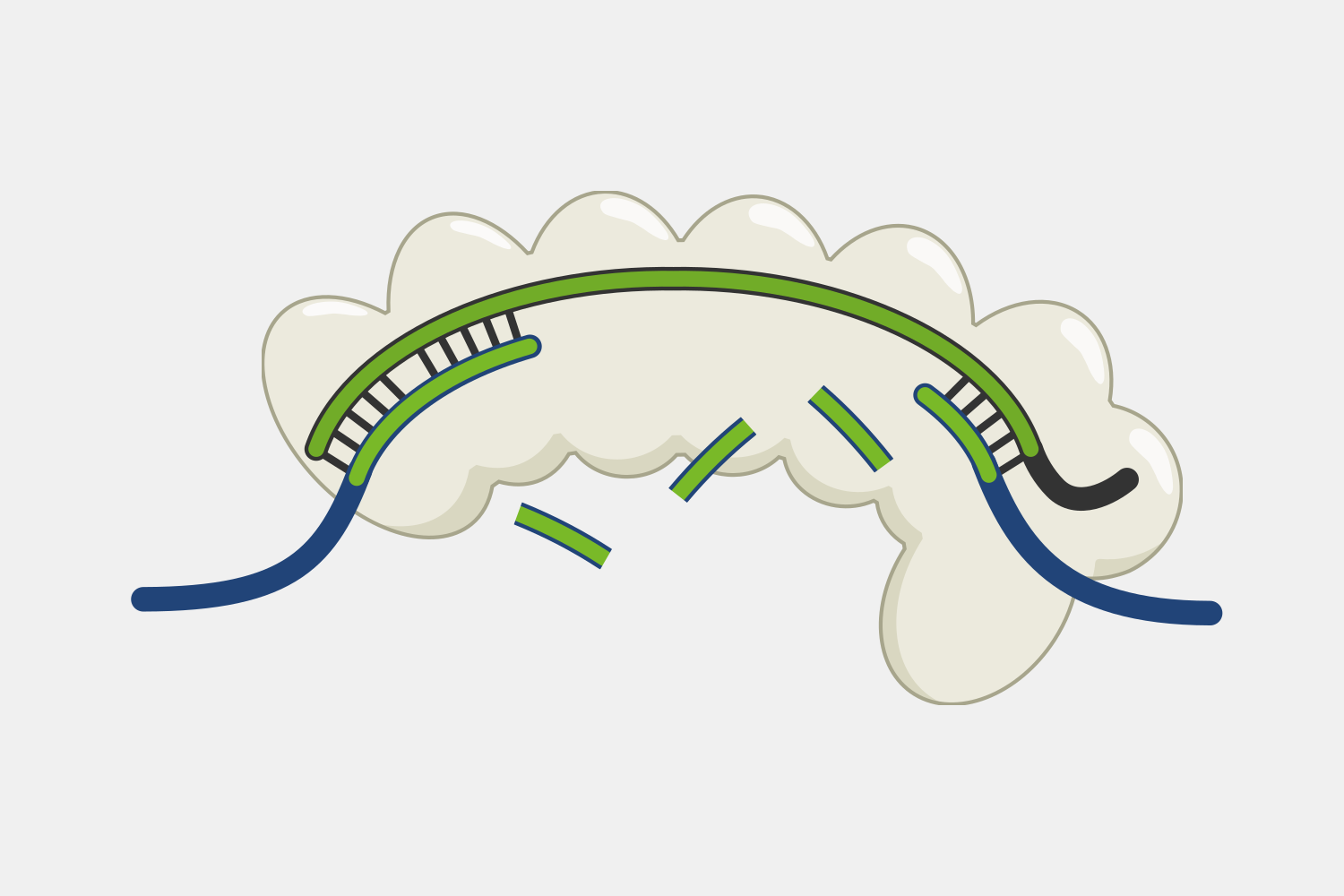
Type III Interference
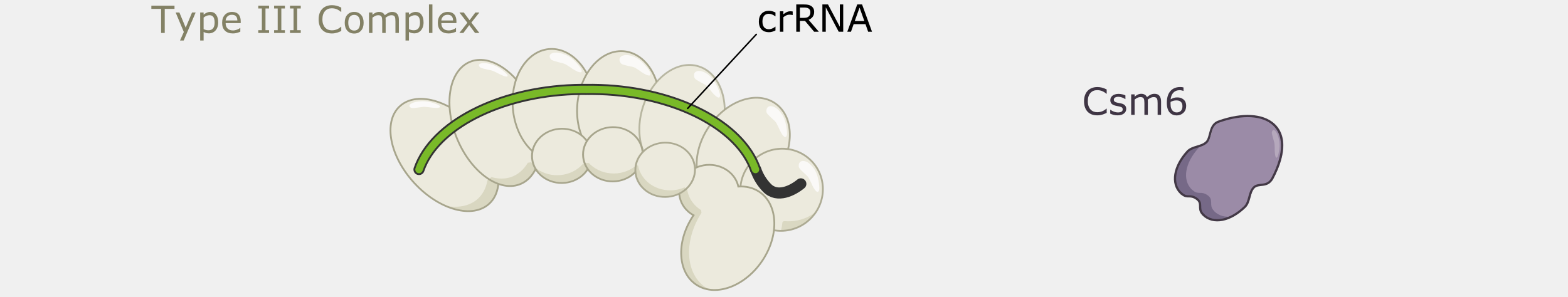
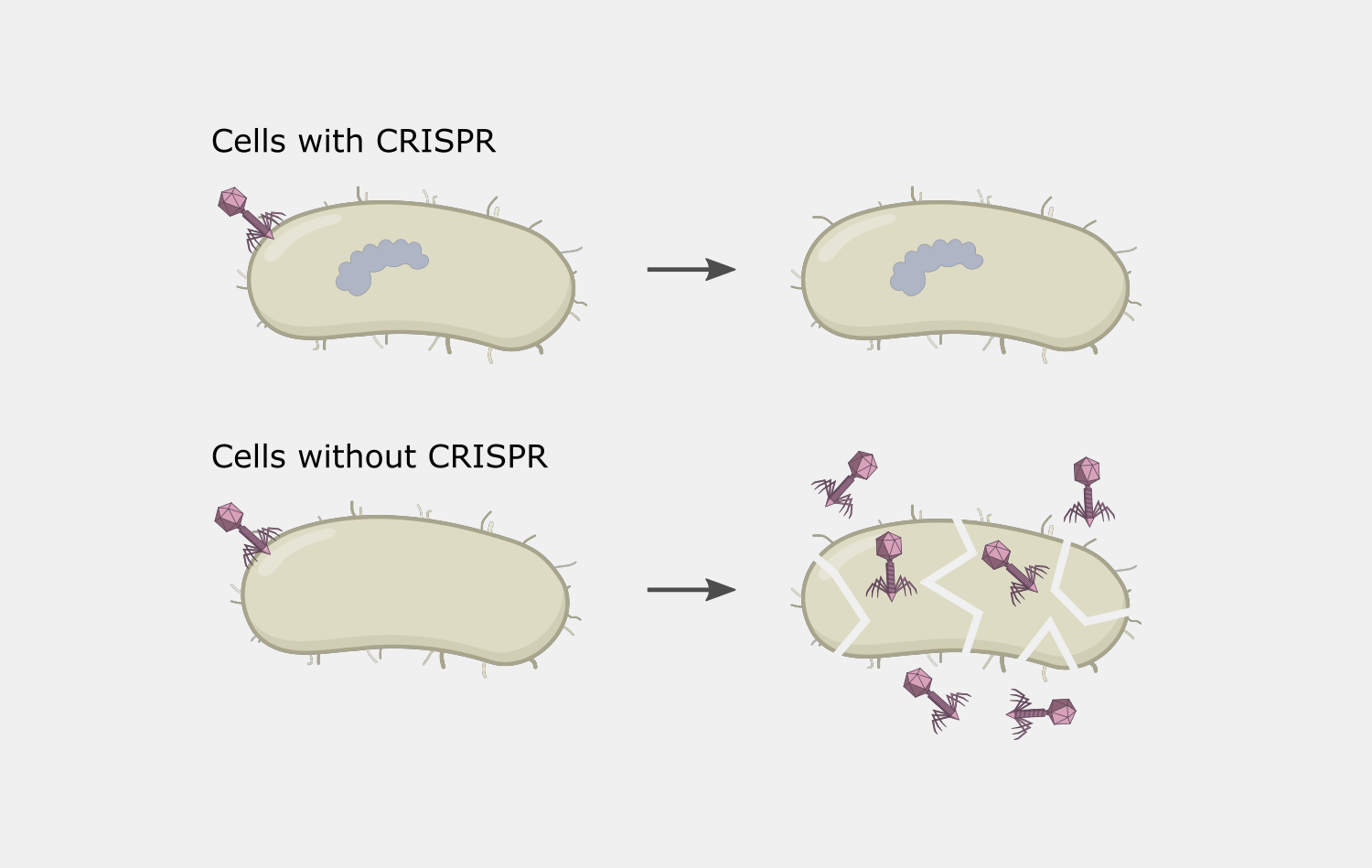
Type III CRISPR systems are unique in that they only cleave phage DNA that is actively being transcribed. They are also capable of triggering cell death if an infection can’t be stopped, a strategy that would kill the host cell but prevent the infection from spreading to nearby cells. Type III CRISPR complexes are made up of multiple proteins assembled on the crRNA and can cleave RNA and DNA. They can also trigger RNA cleavage activity of Csm6 family proteins, which are not part of the complex.
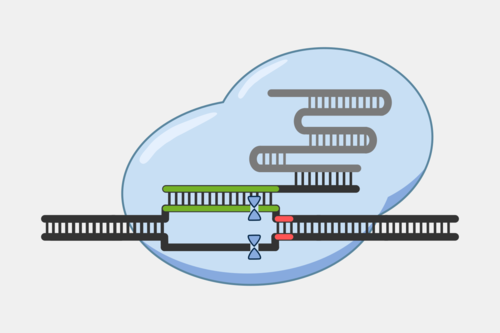
Type III CRISPR complexes target RNA transcripts and use their crRNA to specifically bind complementary sequences through base pairing. This binding triggers several activities within the complex.
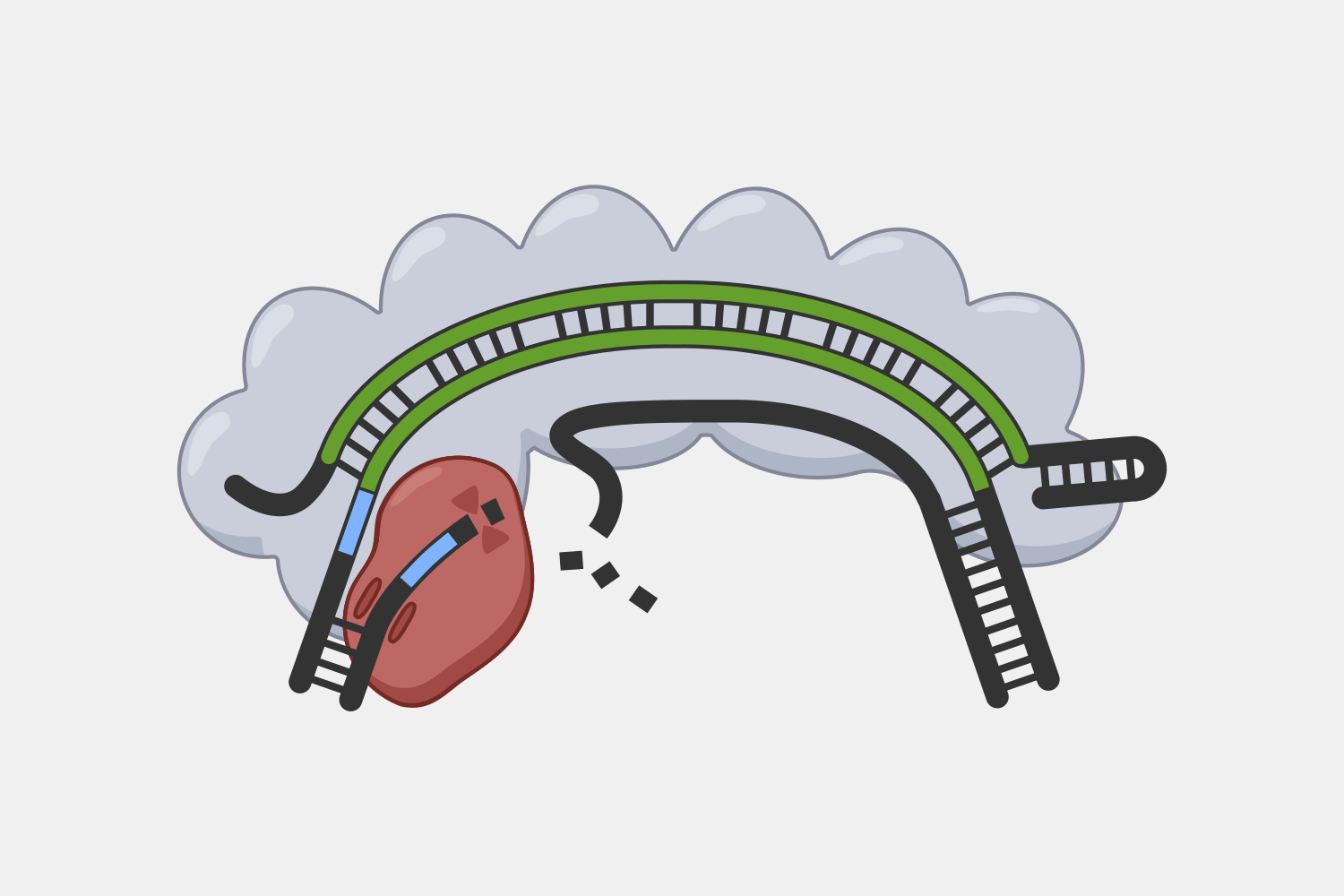
After binding, the complex quickly cleaves single-stranded DNA (ssDNA) within the phage genome, and, at the same time, the complex starts converting ATP into cyclic oligoadenylates (cOAs). cOAs, which act as second messengers, bind to Csm6 proteins and trigger their RNA cleavage activity. Active Csm6 cleavage is not limited to phage RNA, but also cleaves host RNA.
Transcript binding also triggers slow cleavage of the transcript, at multiple sites within the region complementary to the crRNA. Once cleaved, the resulting RNA fragments are released and the Type III complex is deactivated. The slow rate of cleavage prior to RNA release acts as a timer, constraining how long the DNA cleavage and cOA production activities proceed.
The complexity of Type III interference allows for different outcomes depending on the conditions of an infection. If the infection is cleared early, deactivation of DNA cleavage and cOA production activities can limit the potential collateral damage to the host’s own DNA and RNA. If it takes longer to clear the infection, the cumulative amount of active Csm6 may cause cleavage of enough host RNA to lead to cell dormancy, which provides more time to destroy the invading phage. In situations when the phage cannot be destroyed, Csm6 activity could lead to cell death, preventing the infection from spreading to nearby cells.
Protospacer Flanking Sequence (PFS)
Unlike Type I and II systems that target DNA, Type III systems do not use a PAM in the initial steps of interference. However, the DNA cleavage and cOA production activities are blocked if the 5’ handle of the crRNA is complementary to a region of the transcript called the protospacer flanking sequence (PFS). This part of the crRNA is transcribed from the CRISPR array repeat, rather than the spacer. A transcript generated from the cell’s own CRISPR array, transcribed in the incorrect direction, would be recognized by the Type III system, but because the PFS would match the crRNA handle, the transcript cleavage would proceed, but not the collateral DNA and non-specific RNA damage.
Resources
Related Bailey Lab Research
By testing different combinations of target RNA and crRNA sequences, Kaitlin Johnson identified matching requirements for a Thermatoga maritima Type III CRISPR system Target sequence requirements of a type III-B CRISPR-Cas immune system. Johnson, Learn, Estrella and Bailey, 2019, J. Biol. Chem. Also check out our website write-up: What makes a match? For CRISPR type III-B, it depends on location.
Using an in vitro system, Michael Estrella showed that when Thermatoga maritima Type III CRISPR bound a ssRNA that matched the crRNA, the complex cleaved the ssRNA and activated a ssDNA cleavage activity, which cleave even short single-stranded stretches within dsDNA, such as those found in transcription bubbles, resolving ambiguity from previous reports about the system’s nuclease activity RNA activated DNA cleavage by the Type III-B CRISPR-Cas effector complex. Estrella Kuo and Bailey, 2016, Genes & Dev. For more details, check out our post, Solving a targeting puzzle: Type III-B DNA cleavage.
Reviews
Abortive Infection: Bacterial Suicide as an Antiviral Immune Strategy, Lopatina, Tal and Sorek, 2020, Annual Review of Virology
RNA-Targeting CRISPR–Cas Systems and Their Applications, Burmistrz, Krakowski and Krawczyk-Balska, 2020, International Journal of Molecular Sciences
Three New Cs for CRISPR: Collateral, Communicate, Cooperate, Varable and Marraffini, 2019, Trends in Genetics
Shooting the messenger: RNA-targeting CRISPR-Cas systems, Zhu et al, 2018, Bioscience Reports